David Baker is a professor at the University of Washington, a structural biologist, and one of the recipients of the 2024 Nobel Prize in Chemistry. His research focuses on protein folding, protein design, and the structural analysis of biomacromolecules. In 2024 alone, his laboratory published nearly 19 studies in the field of de novo protein design. By integrating artificial intelligence with computational modeling, his team has driven a technological revolution in designing proteins from scratch. They demonstrated how deep learning algorithms, molecular dynamics simulations, and experimental validation can be combined to rapidly and efficiently construct functionally diverse proteins, addressing scientific challenges that have long eluded traditional approaches.
Paper Highlights
The study titled “De novo design of miniprotein agonists and antagonists targeting G protein-coupled receptors”, recently published on bioRxiv, focuses on G protein-coupled receptors (GPCRs), which are central targets in drug discovery and development. Due to their multi-pass transmembrane structure and conformational dynamics, designing protein-based agonists and antagonists for GPCRs has long been a significant challenge. This study presents a comprehensive overview of the design strategy and screening methodology employed.
- This paper combines computational and experimental design approaches to develop GPCR-targeting miniproteins with high affinity, potency, and high selectivity. The method integrates the RFdiffusion, MetaGen, and Receptor-Directed (RD) microscopy-based high-throughput screening.
- Two design strategies were employed:
Motif-guided RF diffusion: Targets specific hotspot regions of the receptor to achieve deep pocket interaction.
MetaGen: Generates diverse designs using a macro-proteome structural scaffold derived from AlphaFold predictions.
- The Receptor-Diversion (RD) screening platform operates directly in human cells, eliminating the need for receptor purification and ensuring compatibility with complex cellular environments. This technique enables efficient screening of up to 100,000 designs.
- The designed miniprotein agonists successfully activated MRGPRX1, a GPCR associated with itch and pain. Two selected binders demonstrated full and partial agonist activity, respectively. Cryo-EM structural data further validated their binding precision and mechanism of action.
- The study also developed antagonists targeting CXCR4, GLP1R, GIPR, GCGR, and CGRPR, with affinities ranging from nanomolar to picomolar levels. Notably, the CGRPR antagonist dC2_049 showed exceptional selectivity and potency.
- Cryo-EM structures confirmed that the designed antagonists block activation by sterically occluding the binding site, offering a novel avenue for GPCR-targeted therapeutics.
- The successful integration of computational design and high-throughput screening offers a promising pathway for developing innovative protein-based therapeutics targeting GPCRs.
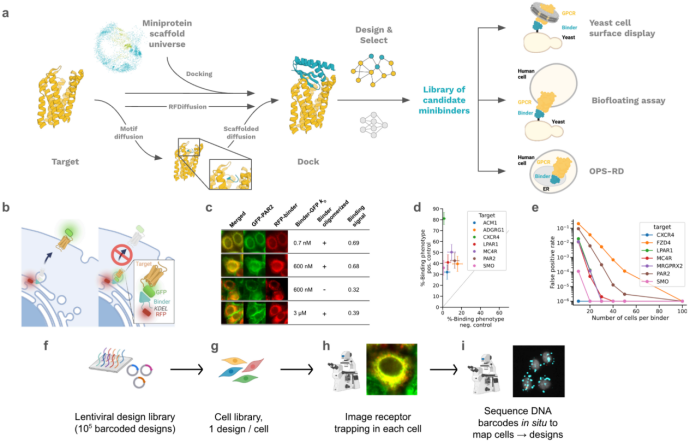
Fig. 1. GPCR binder computational design and screening methods [1]
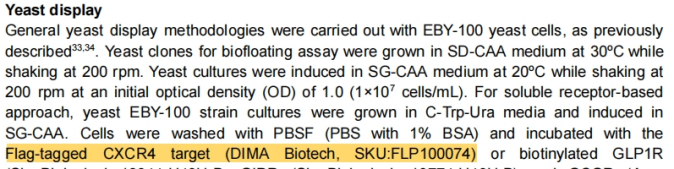
DIMA BIOTECH’s CXCR4-nanodisc Cited in This Paper
DIMA BIOTECH’s CXCR4-nanodisc (FLP100074) was involved in the experimental part of this study, serving in yeast display and screening assays.
DIMA BIOTECH’s nanodisc solution, specifically designed for full-length membrane proteins, addresses key challenges such as poor expression, low yield, poor solubility, and difficult purification of multi-pass transmembrane proteins. It enables the incorporation of transmembrane proteins into a native-like lipid environment, preserving their biological activity. Moreover, downstream experiments can be performed without the need for detergent additives, allowing direct application in cellular assays. The use of a mammalian expression system further ensures proper post-translational modifications, providing additional support for the functional integrity of transmembrane proteins.
To date, DIMA BIOTECH has developed over 500 ready-to-use transmembrane protein products using its nanodisc platform. These products span key target families such as GPCRs and ion channels, with some proteins featuring up to 24 transmembrane domains.
Reference:
[1] https://doi.org/10.1101/2025.03.23.644666