Using multiple recombinant protein fragments from the same target—such as extracellular domains, functional regions, and membrane-proximal segments—has become an essential strategy in therapeutic antibody discovery. This approach improves epitope diversity, enhances screening accuracy, reduces conformational bias, and accelerates the identification of high-value antibodies including blockers, agonists, and internalizing antibodies.
1.Why Full-Length Antigens Often Fall Short?
Developing therapeutic antibodies requires identifying clones that bind specific functional regions of a target protein. However, screening with a full-length antigen alone often introduces several challenges:
- Immunodominance causes strong responses toward exposed, non-functional regions while masking essential functional epitopes.
- Weak immunogenic regions cannot generate robust B-cell responses.
- Complex multi-domain targets may not fold properly when expressed as a single fragment.
- Critical domains may remain inaccessible, limiting the discovery of blocking or agonistic antibodies.
To address these limitations, researchers increasingly rely on domain-specific recombinant protein fragments, which allow selective exposure of functional regions and improve the diversity, accuracy, and druggability of antibody hits.
2.What Are “Different Protein Fragments from the Same Target”?
Protein fragments refer to independently expressed recombinant regions derived from one target protein (e.g., receptor, enzyme, or viral antigen). They are used as standalone antigens to promote epitope-specific immunity or to screen binding interactions more precisely.
Common types of target-specific protein fragments usually include extracellular domain (ECD) fragments, functional domain fragments, mutant vs. wild-type fragments, and membrane-proximal and distal segment. Among of them, extracellular domain (ECD) fragments is ideal for membrane proteins such as immune checkpoint receptors or cytokine receptors (such as domains of CDH17 displayed in the figure 1); Functional domain fragments refer to ligand-binding domains, dimerization interfaces, signaling motifs, etc.; Mutant vs. wild-type fragments are useful for epitope mapping, mechanism-of-action discovery, and affinity maturation; Membrane-proximal and distal segment are different antibody modalities (e.g., ADCs), which may require binding to a specific region—especially membrane-proximal epitopes that promote internalization.
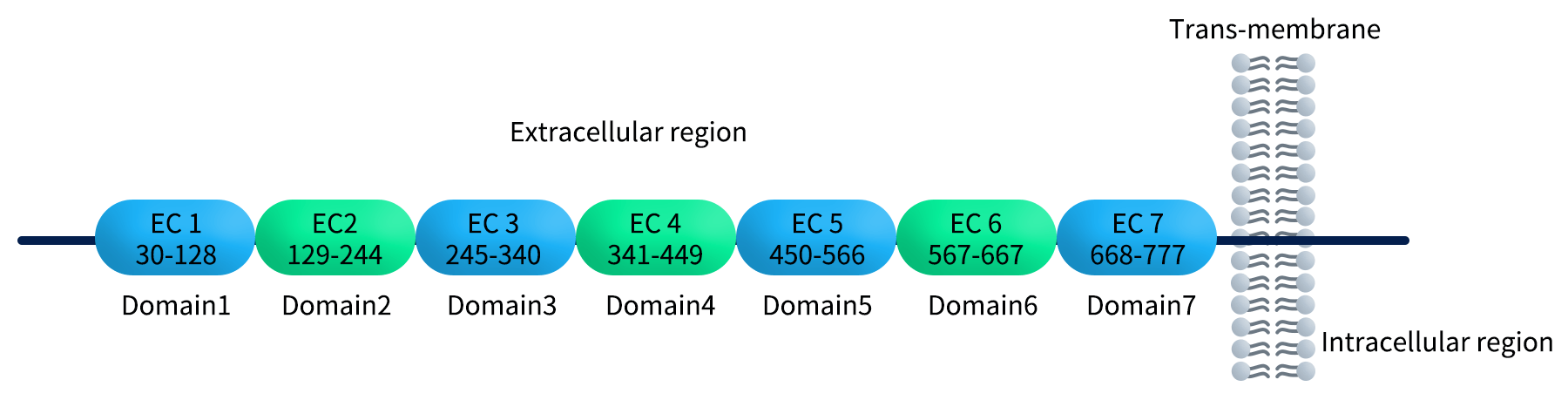
Figure 1. Diverse domains of CDH17
3.Why Antibody Discovery Requires Multiple Protein Fragments?
A single antigen fragment rarely provides complete epitope coverage or accurate folding. Using multiple fragments addresses these gaps and offers several advantages.
3.1 Enhancing Antibody Diversity & Hit Rate
Single fragments expose only a subset of epitopes, restricting B-cell activation.
Multiple fragments:
- broaden epitope coverage
- activate diverse B-cell lineages
- increase hit rate for functional antibodies
- support discovery of blockers, agonists, and internalizing antibodies
3.2 Avoiding Conformational Bias & False Positives
Some fragments misfold or lack intramolecular interactions present in the full-length protein.
Using multiple fragments helps:
- eliminate antibodies targeting misfolded or non-native structures
- confirm structural relevance across fragments
- enrich antibodies closer to the native conformation
3.3 Improving Screening Efficiency & Reducing Development Time
Multi-fragment immunization and alternating-fragment panning strategies allow researchers to:
- cover more epitopes from the first immunization
- reduce the number of booster immunizations
- minimize repeated biopanning rounds
- increase the accuracy of downstream validation
3.4 Supporting Dual-Epitope Antibody Development
For mutation-prone targets, dual-epitope binders such as bispecific antibodies or dual-epitope CAR-T constructs are essential.
Example: BCMA dual-epitope CAR (CARVYKTI®).
Multi-fragment antigen systems make dual-epitope discovery more feasible.
4.Applications of Protein Fragments Across Different Target Classes
Receptors such as EGFR and HER2 have multiple ligand-binding domains and regulatory motifs.
Using domain-specific fragments improves the chance of identifying:
- domain-selective inhibitors
- antibodies recognizing cryptic epitopes
- function-modulating antibodies targeting activation interfaces
4.2 GPCRs & TNFR Family Targets
GPCRs and TNFR family proteins feature complex architectures with multiple extracellular loops and cysteine-rich domains.
- Multi-fragment strategies help:
- avoid antibodies binding non-functional loops
- identify conformation-dependent binders
- obtain antibodies with better mechanism-of-action specificity
4.3 Viral Antigens (e.g., SARS-CoV-2 Spike)
Using RBD, NTD, S1, or engineered mutant variants:
- increases the probability of isolating neutralizing antibodies
- enables epitope mapping and escape-variant assessment
- supports pan-variant antibody development
5.How Protein Fragments Are Used Across Screening Stages
Across the antibody discovery workflow, protein fragments play complementary roles. From immunization to display selection and functional validation, these fragments help researchers capture broader epitope diversity and identify antibodies with true therapeutic potential.
5.1 Immunization – Boosting Immunogenicity
Using mixed or alternating fragments:
- triggers broader B-cell activation
- enhances immune recognition of weak yet functional epitopes
- reduces immunodominance bias
5.2 Display Screening – Improving Panning Specificity
Phage, yeast, or mammalian display can incorporate fragment-swapping panning:
Round 1: fragment A
Round 2: fragment B
Round 3: full-length ECD or mutant fragment C
This prevents enrichment of antibodies binding irrelevant or misfolded regions.
5.3 Secondary Validation – Cross-Fragment Epitope Confirmation
Using multiple fragments in ELISA, SPR, BLI, or flow cytometry enables:
- epitope localization
- filtering non-specific binders
- validating functional region recognition
6.Production & Quality Control of Protein Fragments
High-quality fragment production requires native folding, correct disulfide bonding, and proper post-translational modifications—often difficult in bacterial, yeast, or insect systems.
For therapeutic antibody discovery, mammalian expression systems are preferred because they:
- ensure correct folding and glycosylation
- maintain native structural integrity
- deliver superior biological activity
- produce high-purity, reproducible antigens for screening
DIMA BIOTECH has developed a comprehensive library of domain-specific protein fragments for critical therapeutic targets, expressed using high-yield mammalian expression platforms.
Representative multi-fragment targets include:
Click for target details